DOG1
DOG1 is the key regulator of dormancy strength. DOG1 expression is extensively regulated. We showed that DOG1 is regulated by several lncRNA.
An antisense transcript 1GOD transcribed from within DOG1 locus supresses dormancy by inhibiting DOG1 eaxpression in cis. DOG1 antisense is itself negatively regulated by ABA in seedlings and by DOG1 alternative polyA site selection in seeds.
PUPPIES are a group of sense lncRNA transcripts transcribed from DOG1 promoter that activate DOG1 expression in freshly harvested stratified seeds in response to presence of salt, delaying germination. PUPPIES activate DOG1 expression by enhanced pausing of RNA polymerase II, slower transcription and higher transcriptional burst size.
Further reading:
Montez, EMBO 2023
Fedak and Palusinska, PNAS 2016
Yatusevich, EMBO Rep. 2017
Archacki, NAR 2017
Kowalczyk Mol Plant. 2017
Dormancy
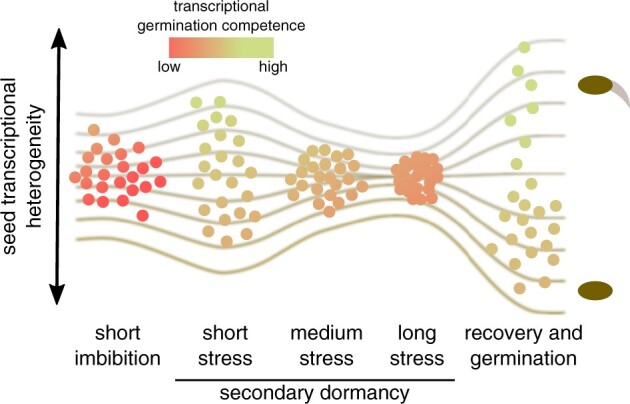
Seed dormancy strength is usually analysed as percentage of seeds in a seed lot that germinate or postpone germination. We are analysing the variability among individual seeds focusing on the transcriptional variability between individual seeds. To do this we developed a single seed RNA sequencing method that allows to interrogate transcriptomes of hundreds of seeds at the same time. This allowed us to describe transcriptional variability among individual seeds that could underlie the physiological variability in dormancy strength among seeds from an individual seed lot.
Further reading:
Krzysztoń, Plant Physiology 2022
Krzysztoń, Sacharowski, Plant Com. 2024
Transcriptional and posttranscriptional gene expression regulation during seed germination

Seeds adopt a unique strategy, pre-producing numerous mRNAs during maturation, and storing them for subsequent protein synthesis during germination. This requires distinctive posttranscriptional regulation that involves the activation and removal of specific mRNAs, guided by specialized enzymatic machinery upon water uptake. We are investigating how Arabidopsis thaliana seeds regulate germination under normal and stress conditions through posttranscriptional mechanisms.
Dormancy and DOG1
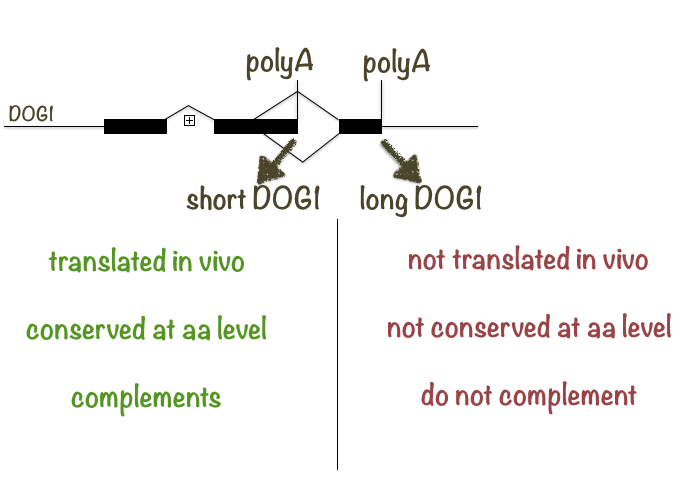
We found that alternative polyadenylation results in production of two DOG1 transcripts: a shorter two exonic short DOG1(shDOG1) and longer three exonic long DOG1 (lgDOG1). Our work showed that the shDOG1 transcript is evolutionary conserved at the level of amino acids, translated and sufficient to complement dog1 mutant.
Further reading:
Cyrek and Fedak Plant Phys 2016 click for PDF
DOG1 alternative splicing
DOG1 alternative splicing. Second intron of DOG1 is subject to alternative splicing, generating four different isoforms. By studying the regulation of this alternative splicing we described a tight coupling between transcription and alternative splice site selection. We showed that mutations in transcription elongation factor TFIIS lead to selection of proximal splice sites not only on DOG1 but also at majority of tested genome wide targets - suggesting existence of a kinetic coupling between PolII elongation and splicing in plants. We have also shown that alternative splice sites are active players in PolII elongation control. We hypothesized that alternative splicing may locally pause the elongating PolII using a chromatin based mechanism that is centered around the spliceosomal disassembly factor NTR1.
We have also helped to describe a mechanism where alternative splicing control by light is mediated by changes in Pol II speed that is dependent on the TFIIS elongation factor.
Further reading
Dolata and Guo, EMBO J 2015 click for PDF
Brzyzek Transcription 2015 click for PDF
Godoy Herz Mol Cell 2019 click for PDF
DOG1 antisense regulation
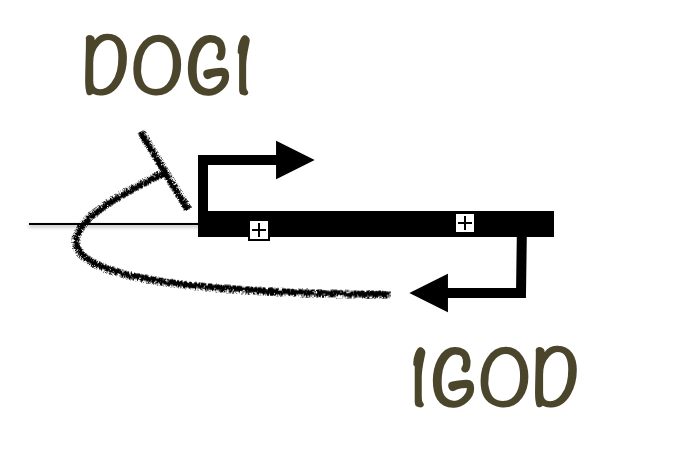
Transcription generates an extensive array of non-protein-coding RNA (ncRNA), the functional significance of which is mostly unknown. Studies in yeast, humans, drosophila, bacteria and plants have shown that the majority of nucleotides in these genomes are transcribed, generating a mixture of long and short RNAs, most of which are non-coding. This non-coding transcription changes in a complex manner during development and in response to environmental signals. One specific class of ncRNA are antisense transcripts.
We have recently characterized an antisense transcript originating close to DOG1 proximal (main) termination site that strongly inhibits dormancy strength and DOG1 expression. We further showed that this transcript acts in cis as it is unable to silence DOG1 transcribed from a second allele.
We also showed that in mature plants DOG1 antisense acts as a sensor for a plant hormone ABA allowing the mature plant to upregulate DOG1 expression in response to drought. This allowed us to characterise DOG1 function outside seed in conferring increased drought tolerance.
Further reading
Fedak and Palusinska, PNAS 2016 click for PDF
Yatusevich, EMBO Rep. click for PDF
For list of all papers from the lab please click here